T4K3.news
Kidney Proteomics Maps New Targets for CKM health
New kidney proteomics work links tissue proteins to cardio-kidney-metabolic traits and introduces a kidney pQTL map to guide future therapies.
A large kidney proteomics study links tissue proteins to cardio-kidney-metabolic traits and introduces a kidney pQTL map to guide future therapies.
Kidney Proteomics Reveals New Targets for Cardio-Kidney-Metabolic Health
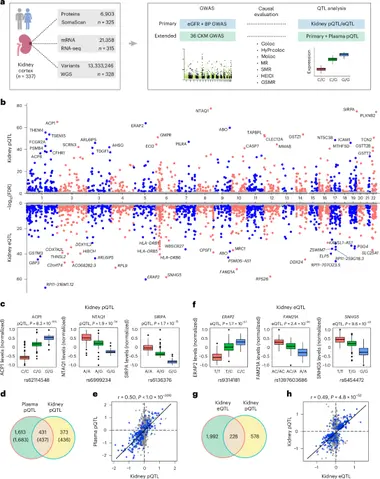
Ndumele and colleagues analyzed 337 human kidney samples with whole-genome sequencing, RNA sequencing, and deep proteomics. They built a kidney protein quantitative trait loci (pQTL) resource and, together with 36 cardio-kidney-metabolic GWAS, used Bayesian co-localization and Mendelian randomization to prioritize 89 proteins for CKM traits. Among the standout signals are ANGPTL3, implicated in serum lipid levels and kidney function, and CHMP1A, linked to kidney function and hypertension.
The team also created a publicly accessible database that integrates genetic, transcript, and protein data. The study argues that kidney proteomics captures mechanisms not visible in gene expression profiles or blood proteomes, and that convergent signals across data layers point to actionable pathways. The authors say the work lays a foundation for targeted therapies but emphasize the need for functional validation and diverse cohorts before clinical use.
Key Takeaways
"This work shows tissue proteomics can reveal mechanisms genetics misses."
Describes the added value of proteomics beyond genetics.
"Angptl3 in the kidney could reshape how we think about lipid and kidney health."
Relates to the ANGPTL3 finding.
"A kidney pQTL map is the missing piece in cardio-kidney-metabolic disease."
Highlights the novelty of the dataset.
"A public proteomics resource may speed up precision medicine for cardio-kidney-metabolic health."
Editorial closing thought on impact.
The work highlights how proteomics can fill gaps left by transcriptomics and plasma data. By mapping pQTLs in kidney tissue, scientists can connect genetic variation to protein abundance and disease traits in a tissue-specific way. The finding that ANGPTL3 in kidney might influence both lipids and kidney function points to a new cross-talk pathway. But the results are correlational and require lab experiments to test causality.
Yet translating such maps into therapies faces hurdles: replication in diverse populations, understanding tissue context, and moving from association to mechanism. The public database is a valuable resource, but clinicians need clear validation and safe, ethical deployment. The study nudges the field toward precision medicine for CKM health, but it also calls for careful, patient-centered development.
Highlights
- Kidney proteins unlock what genes alone cannot
- Public proteome maps change drug target discovery
- Angptl3 in kidney links lipids to function
- Proteomics adds depth to cardio-kidney insight
Future studies will test whether these kidney signals translate into real patient benefits.
Enjoyed this? Let your friends know!
Related News

Midlife aging shift identified

BioNTech shows promise in cancer therapy
Measles hotspots rise as vaccination gaps widen

Natural pain relief explored by experts

Visceral fat linked to faster heart ageing
Study Links Gut Health to Chronic Fatigue and Long COVID
Estrogen protects kidneys from ferroptosis

Major Study Identifies Age 50 as Crucial Aging Milestone
