T4K3.news
ALPACA reveals lineage driven copy-number evolution in lung cancer
A new method maps how copy-number changes evolve across tumour lineages and links diversity to survival
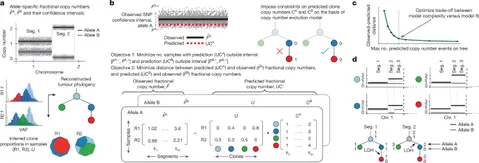
ALPACA infers clone specific copy number evolution from multi sample bulk sequencing and links higher copy number diversity to worse outcomes in lung cancer.
Clone copy number diversity is linked to survival in lung cancer
ALPACA is a new algorithm that infers allele specific copy number profiles for every tumour clone using bulk sequencing data from multiple samples. It takes as input allele specific fractional copy numbers per genomic segment, a fixed number of clones with their proportions in each sample, and a fixed tumour phylogenetic tree. The method then models copy-number evolution along the tree with irreversibility constraints for loss of an allele and a cap on changes along any path, before solving a mixed integer linear program to estimate the actual copy numbers for each clone and each segment.
In the TRACERx lung cancer cohort ALPACA uncovers events in minor clones that standard approaches can miss, including loss of heterozygosity and amplifications. The study reports higher chromosomal instability in metastasis seeding clones and enrichment for losses in tumour suppressor genes and amplification of CCND1. It also finds elevated SCNA rates in polyclonal and extrathoracic metastases, and links greater clone level copy-number diversity to shorter disease free survival. The authors benchmark ALPACA against existing tools on simulated data, showing improved accuracy and scalability, and apply the method to single cell derived data for validation.
Key Takeaways
"A tree guides the copy-number story not a single snapshot"
Emphasizes the tree based approach over snapshot views
"Minor clones carry gains and losses that escape ordinary methods"
Pointing to the value of clonal resolution
"Higher clone copy-number diversity associates with reduced disease-free survival"
Links genomic diversity to patient outcomes
"ALPACA turns bulk data into lineage aware copy-number maps"
Summarizes the method’s key strength
ALPACA advances copy-number analysis by turning bulk data into a lineage aware map of events. By fixing clone proportions and using a phylogeny derived from SNVs, it reduces a complex deconvolution into a more tractable linear optimization. This lineage grounded view helps separate trunk changes from branch changes and clarifies when copy-number events may drive metastasis.
But the approach relies on accurate input trees and clone proportions, which can be uncertain in practice. The irreversibility assumption for LOH and the elbow based model selection may limit applicability in noisy data. Still, the linkage between higher clone diversity and worse outcomes offers a possible biomarker direction and a rationale for tracking copy-number evolution in trials and future therapies.
Highlights
- A tree guides the copy-number story not a single snapshot
- Minor clones carry gains and losses that escape ordinary methods
- Higher clone diversity tracks with shorter disease free survival
- ALPACA turns bulk data into lineage aware copy-number maps
As methods mature, lineage based maps of copy-number evolution could guide prognostic tools and targeted research gaps.
Enjoyed this? Let your friends know!
Related News

Jessie J readmitted to hospital after cancer surgery
Common viruses may reactivate dormant cancer cells

Study links processed food to lung cancer risk

Warning over sucralose impact on cancer treatments

Shark DNA puzzles deepen
Proteins Extend the Human Evolution Narrative Beyond DNA

Link between ultra processed foods and lung cancer risk identified

Jessie J hospitalized due to health issues
